Bio x ML hackathon: our 3rd-place winning project
Generative DNA design for plasmid vector engineering
Over the past week, hundreds of hackers and researchers came together from across the globe to participate in Evolved 2024, a Bio x ML hackathon hosted by Lux Capital, Evolutionary Scale, and Enveda Biosciences.
The timing was ripe, as just two weeks ago the Nobel Prize in Chemistry was awarded to David Baker, Demis Hassabis and John Jumper for computational protein design and protein structure prediction. The momentum in the field of computational biology is palpable, and this recognition added some great tailwind to the competition.
We had prizes and resources contributed by Evolutionary Scale, Enveda, Recursion, Latch, Ginkgo, Scale Medicine, Adaptyv, SynBioBeta, Polaris, Nvidia, OpenAI, AWS, RunPod, Modal, Together AI and DigitalOcean. Judges represented these resource providers, as well as folks from DARPA, Story Health, Sphinx Bio and Microsoft. The diverse range of business models—from cloud compute providers to drug discovery companies—spanning early-stage startups to billion-dollar enterprises, highlights the growing excitement at the intersection of biology and computing.
In late September, participants from around the world gathered on a Discord server to pitch project ideas and form teams. Project proposals were submitted on September 28, and 20 finalist teams were selected. The hackathon officially began on October 10, and we spent the week building and experimenting. Final projects were submitted on October 20, followed by the award ceremony that same day.
To our surprise and excitement, our team, GenPlasmid, won 3rd Place and the Polaris New Artifact Challenge!
I look forward to gathering more details about the 1st and 2nd place winners, but for now I want to share the details of our project. This is a compilation of our DevPost submission and slides we put together for our video submission. Find the details on team and project outputs at the end of the post.
Generative DNA design for plasmid vector engineering
Inspiration
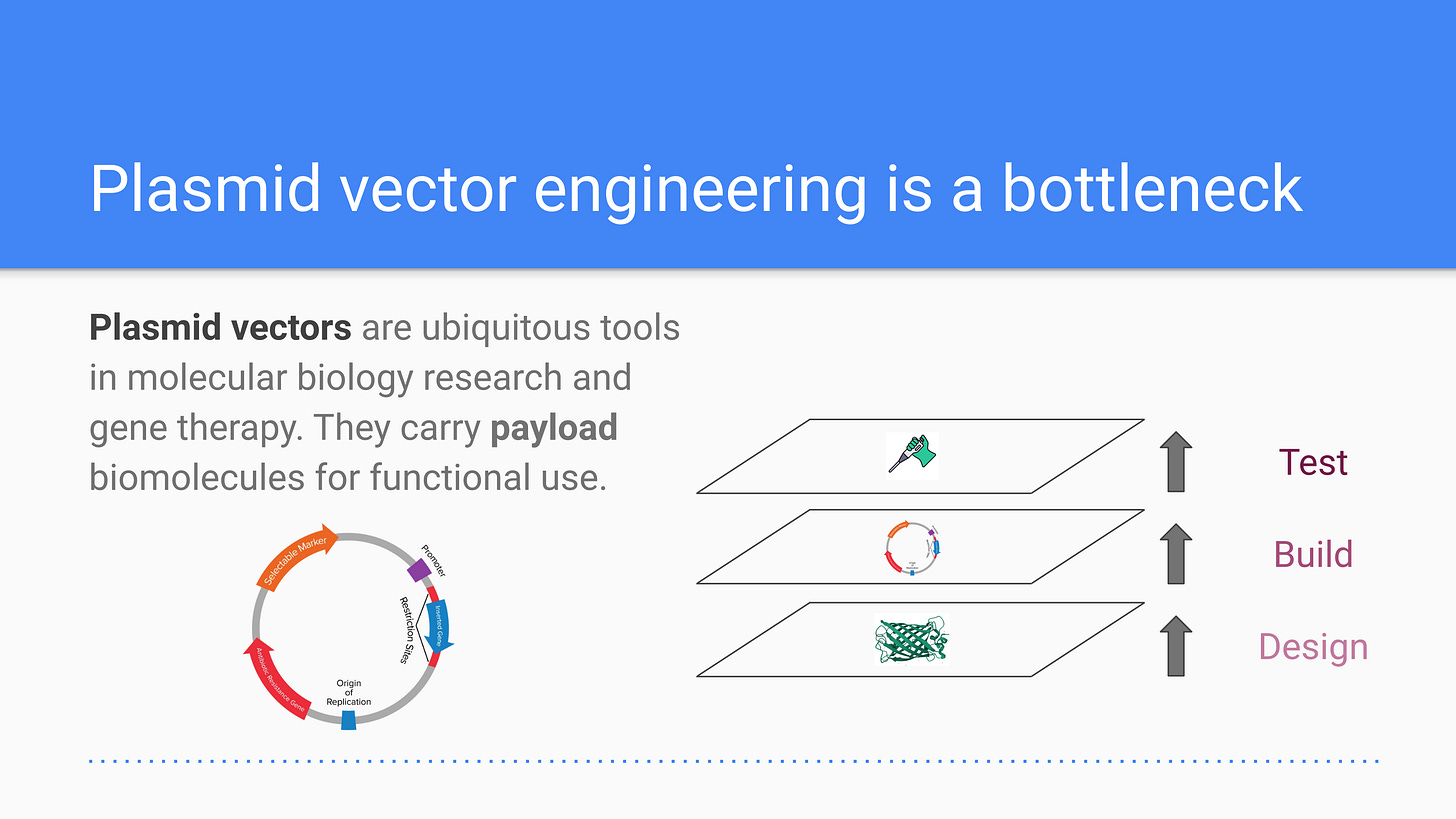
Plasmids are circular DNA molecules capable of replicating independently within a host cell and are essential tools in molecular biology. They play a critical role in gene cloning, protein expression, and reporter assays, making them indispensable for research, synthetic biology, biomanufacturing, and therapeutic development. However, traditional plasmid design is labor-intensive, requiring multiple rounds of experimental validation to achieve optimal gene expression, stability, safety, and host compatibility.
As access to computationally-designed proteins becomes broadly accessible, functional protein expression and testing are emerging as significant bottlenecks. Moreover, specific research questions almost always require highly customized experimental systems for validation. Improving plasmid design can bridge the gap between computational protein design and physical testing, enabling faster and more effective wet-lab validation. GenPlasmid was inspired by the need to enhance plasmid engineering, adding a critical component to the design, build, and test stack in generative biology.
What it does
GenPlasmid automates the design of plasmid components. We validated a use case by applying in silico mutagenesis to generate novel promoters for enhanced expression of YFP.
How we built it
First, we compiled a new dataset, OpenPlasmid, consisting of ~150k engineered plasmids from Addgene. We then finetuned gLM2, a mixed-modality DNA and protein sequence model, and evaluated the learned plasmid representations using a new benchmark, showing improvements over several baselines. Finally, we applied in silico mutagenesis to design novel promoters for YFP expression and computationally evaluated these designs through a robust oracle model.
Results
As part of this project, we curated OpenPlasmid, a new dataset featuring ~150k plasmid sequences sourced from Addgene.
Key features of OpenPlasmid include:
Metadata of each plasmid, providing context on the plasmid’s use and design.
Fully annotated GenBank sequences, which detail the plasmid’s genetic components
We have shared the code and the dataset for public use online.
Next, we built a model for generative plasmid design. We fine-tuned gLM2, from Tatta Bio, on our OpenPlasmid dataset. The goal was to improve the model’s ability to capture meaningful plasmid representations.
To evaluate the effectiveness of the fine-tuning, we designed tests that assess how well the plasmid embeddings reflect key genomic features:
Coding sequence features (CDS-curated-features)
Payload gene features (Entrez-curated-features)
Our fine-tuned model outperformed:
Simple one-hot encoding
A (super recent!) alternative approach, PlasmidGPT1
The original gLM2 model
These results show that our approach is superior in capturing plasmid structure and functionality and motivated its use for promoter design.
To design new promoters, we used an in silico mutagenesis approach. Here’s how it worked:
We started by sampling “seed” sequences from a dataset of promoter sequences. This dataset is from the Random Promoter Sequence DREAM Challenge.
We then used our model to predict randomly masked nucleotides in the seed sequences.
Using this process, we can rapidly generate thousands of promoter sequences.
To evaluate our generated promoters, we used a promoter sequence-to-expression model from the DREAM challenge as an oracle.
We found that our generated promoters are predicted to significantly increase expression levels compared to in vitro promoters.
We also iteratively sampled seed sequences from different percentiles of the in vitro expression distribution. The expression distributions of these generated sequences showed significant improvements in expression.
These results highlight the potential of using in silico mutagenesis to iteratively optimize promoter sequences for payload expression.
So how does this work? Why does a model filling in nucleotides result in promoter sequences predicted to enhance expression? This is a key detail we didn’t cover in the final pitch. The genomic language model likely succeeds because it integrates two key constraints:
Evolutionary pressures from gLM2 pretraining on a large metagenomic corpus
Engineered plasmid requirements from fine-tuning on the OpenPlasmid dataset
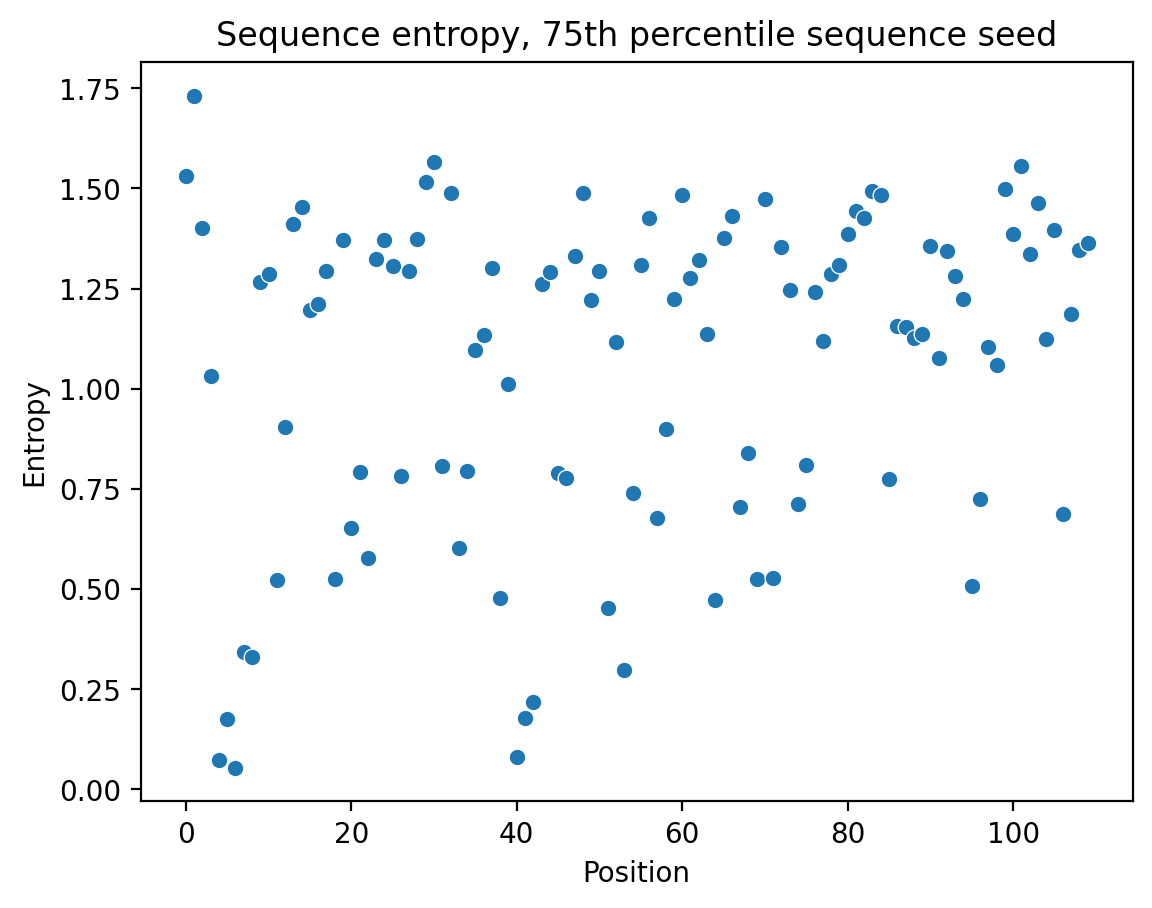
By predicting nucleotides that reflect biologically relevant patterns, the model generates sequences that are likely functional, rather than arbitrary. We can observe this by analyzing nucleotide entropy at each position in the designed promoter sequences—low entropy suggests critical nucleotides, reflecting their evolutionary conservation.
Challenges we ran into
Collecting and organizing the OpenPlasmid dataset was a significant challenge. Initially, we aimed to evaluate the design characteristics of promoters in the context of novel protein sequence variants. We designed thousands of likely functional YFP variants using ESM3 and used these variants for the conditional generation of novel promoters. However, we lacked a robust evaluation framework to report conclusive results, particularly within the time constraints of the hackathon. We plan to continue this work.
Accomplishments that we're proud of
OpenPlasmid: a new, publicly available dataset of 150k annotated plasmid designs.
A state-of-the-art finetuned, mixed-modality genomic language model for plasmid design, with new benchmark evaluations.
Improved promoter design through in silico mutagenesis, and a new benchmark evaluation.
What we learned
Model and task benchmarks are at a premium for validating new methodologies in generative biodesign. Overall, ML in biology is rapidly democratizing, as evidenced by the ability of a globally distributed team to collaborate. We are excited about how access to these tools will compound progress in the field.
What's next for GenPlasmid
Next, we aim to enhance GenPlasmid’s capabilities by incorporating more structured plasmid annotations and developing new frameworks to evaluate tasks of interest, such as delivery and host compatibility. We also want to more directly test the feasibility of “in silico directed mutagenesis.” Ultimately, we want to make GenPlasmid an accessible resource for labs and researchers, enabling plasmid engineering and facilitating data sharing on the success of computational designs.
Team
Karm Aggarwal
Leon Koh
Project resources
Of note, PlasmidGPT was published just a few days after our project was accepted as a finalist! I’m glad other folks see the value in this direction.